PhyloNet
Download Version 3.8.2PhyloNet is a tool designed mainly for analyzing, reconstructing, and evaluating reticulate (or non-treelike) evolutionary relationships, generally known as phylogenetic networks. Various methods that we have developed make use of techniques and tools from the domain of phylogenetic trees, and hence the PhyloNet package includes several tools for phylogenetic tree analysis.
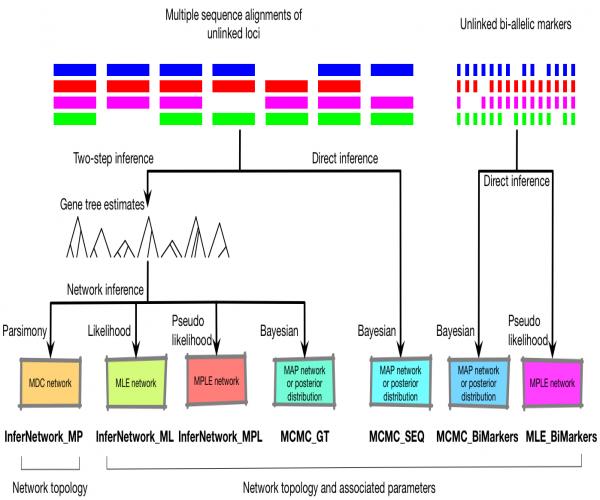
- Commands List (see the figure at the bottom of this page for a summary of the available inference methods)
- Talk about PhyloNet (June 4, 2018, by Luay Nakhleh at the SSB Standalone Meeting in Ohio)
- Tutorial (3 January 2020) at the 2020 Phylogenomics Software Symposium held in conjunction with the SSB Standalone Meeting in Gainesville, Florida.
If you use PhyloNet, please cite
Wen, Y. Yu, J. Zhu, L. Nakhleh (2018) Inferring phylogenetic networks using PhyloNet, Systematic Biology 67(4): 735-740.
C. Than, D. Ruths, L. Nakhleh (2008) PhyloNet: A software package for analyzing and reconstructing reticulate evolutionary histories, BMC Bioinformatics 9:322.
as well as other the publications relevant to the method being used.
A special thank you:
Development of the various functionalities in PhyloNet has been generously supported by DOE (DE-FG02-06ER25734), NIH (R01LM009494), and NSF (CCF-0622037, DBI-1062463, CCF-1302179, CCF-1514177) grants and Sloan and Guggenheim fellowships to Luay Nakhleh.
If you use PhyloNet, kindly fill out this form that allows us to have a better idea who uses our software!